
Blase Mapping Result
MappingResult-class.RdCreated by map_best_bin()
Slots
bulk_nameThe name of the bulk sample being mapped.
best_binThe bin that best matched the bulk sample.
best_correlationThe spearman's rho that the test geneset had between the winning bin and the bulk.
top_2_distanceThe absolute difference between the best and second best mapping buckets. Higher indicates a less doubtful mapping.
confident_mappingTRUE when the mapped bin's lower bound is higher than the maximum upper bound of the other bins.
historyA dataframe of the correlation score and confidence bounds for each bin. Access with
mapping_history()bootstrap_iterationsThe number of iterations used during the bootstrap.
Examples
counts_matrix <- matrix(
c(seq_len(120) / 10, seq_len(120) / 5),
ncol = 48, nrow = 5
)
sce <- SingleCellExperiment::SingleCellExperiment(assays = list(
normcounts = counts_matrix, logcounts = log(counts_matrix)
))
colnames(sce) <- seq_len(48)
rownames(sce) <- as.character(seq_len(5))
sce$cell_type <- c(rep("celltype_1", 24), rep("celltype_2", 24))
sce$pseudotime <- seq_len(48) - 1
blase_data <- as.BlaseData(sce, pseudotime_slot = "pseudotime", n_bins = 4)
genes(blase_data) <- as.character(seq_len(5))
bulk_counts <- matrix(seq_len(15) * 10, ncol = 3, nrow = 5)
colnames(bulk_counts) <- c("A", "B", "C")
rownames(bulk_counts) <- as.character(seq_len(5))
# Map to bin
result <- map_best_bin(blase_data, "B", bulk_counts)
result
#> MappingResult for 'B': best_bin=1 correlation=1 top_2_distance=0
#> Confident Result: FALSE (next max upper 1 )
#> with history for scores against 4 bins
#> Bootstrapped with 200 iterations
# Map all bulks to bin
results <- map_all_best_bins(blase_data, bulk_counts)
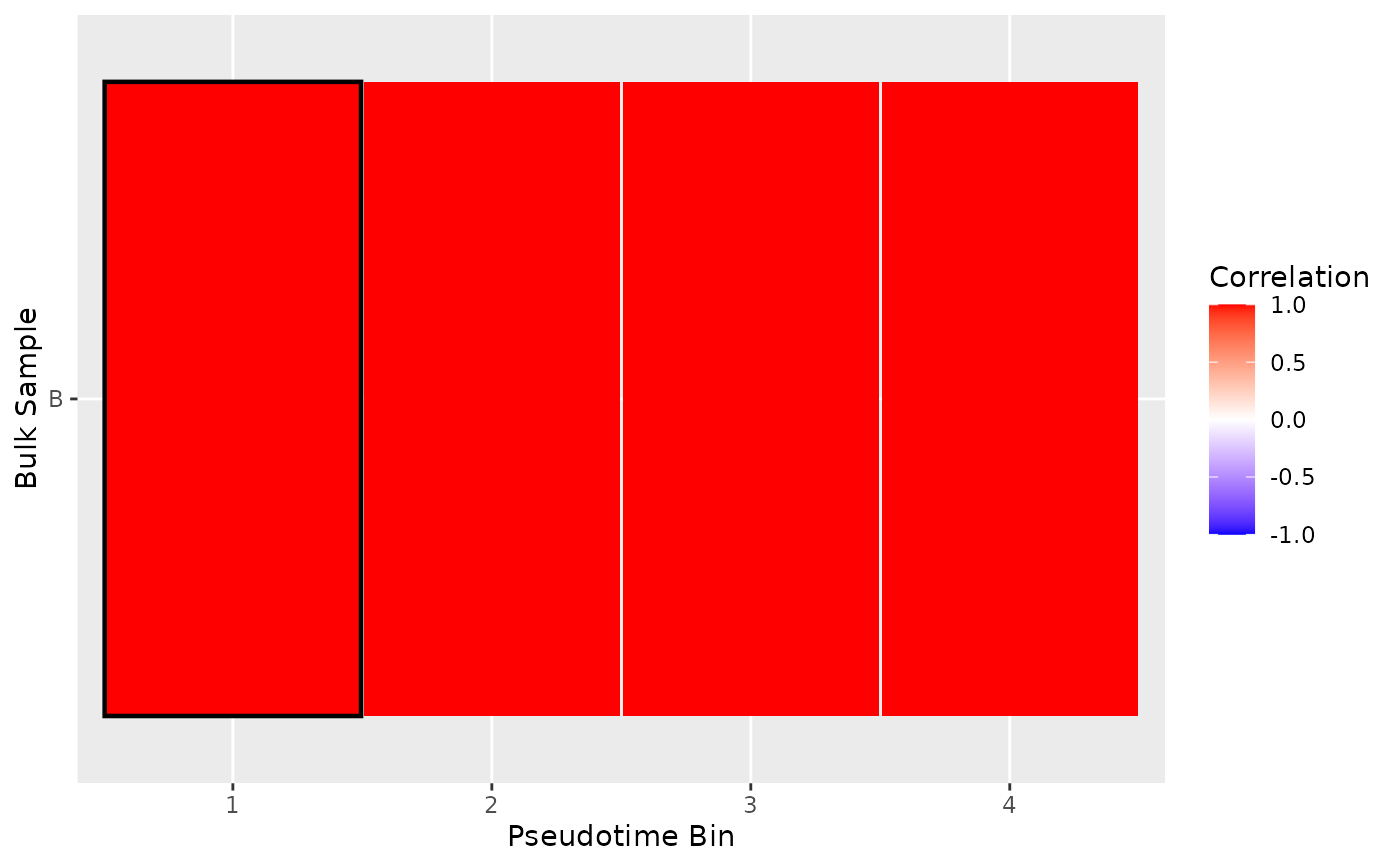
# Plot Heatmap
plot_mapping_result_heatmap(list(result))
 # Plot Correlation
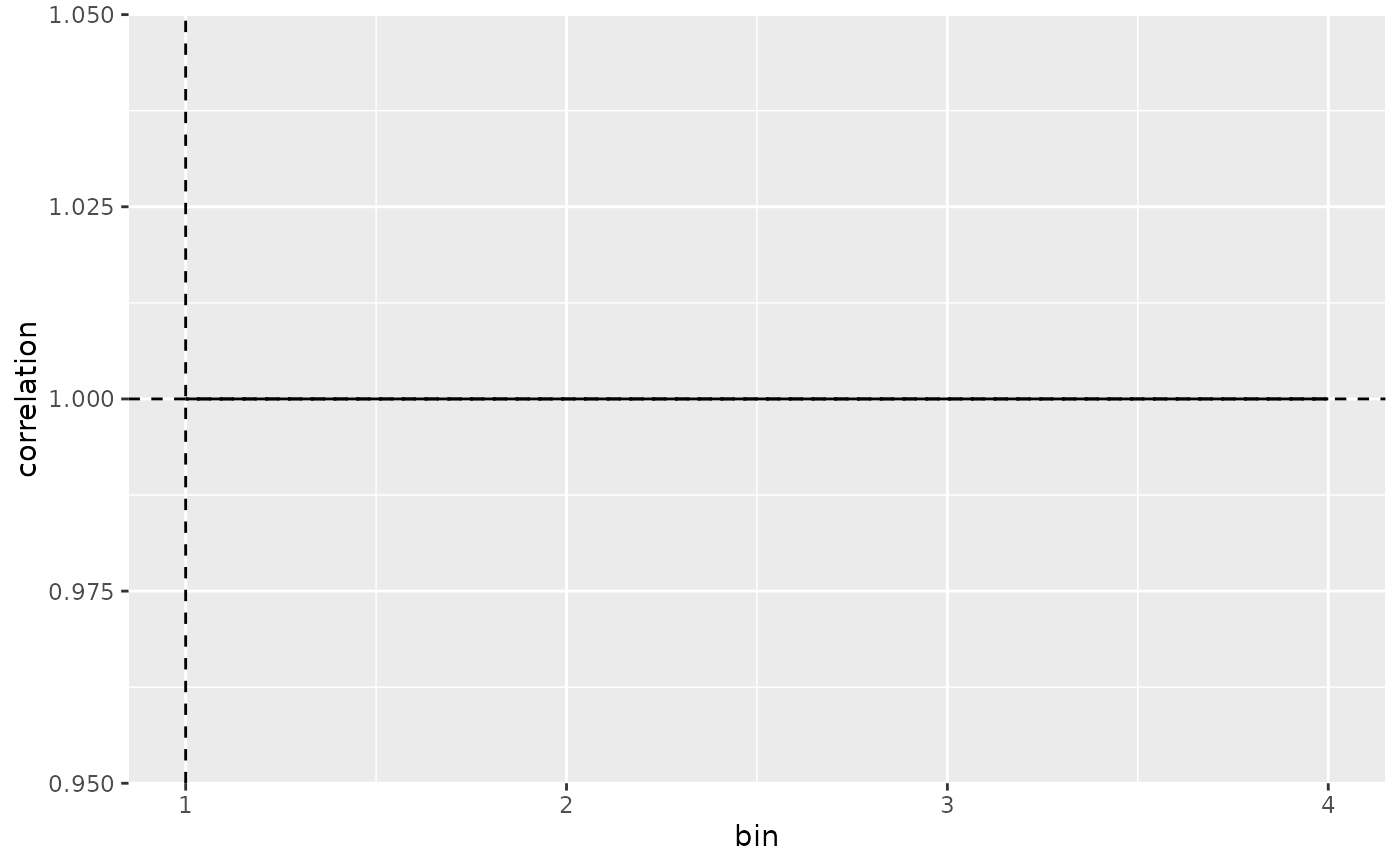
plot_mapping_result_corr(result)
# Plot Correlation
plot_mapping_result_corr(result)
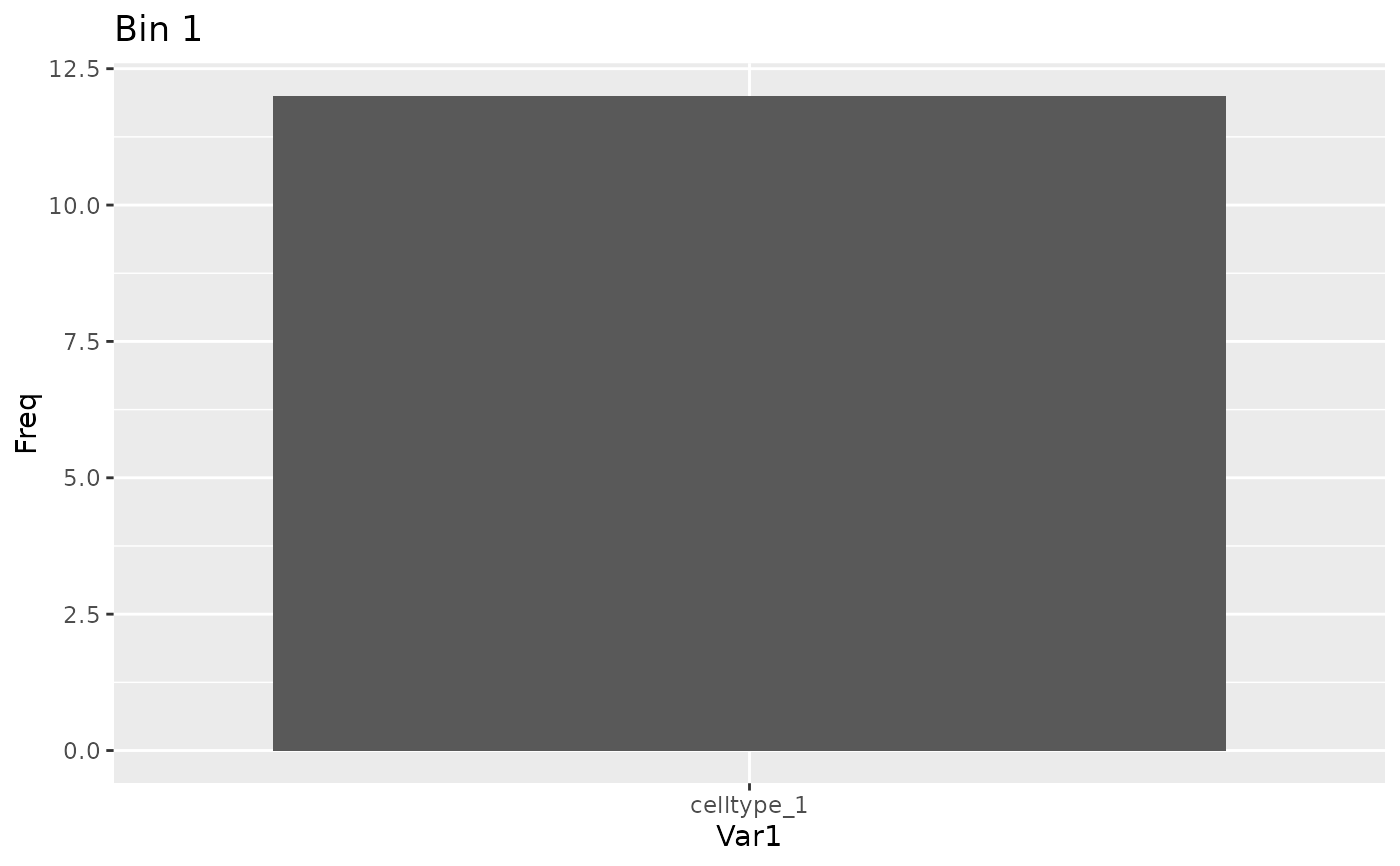
 # Plot populations
sce <- assign_pseudotime_bins(
sce,
pseudotime_slot = "pseudotime", n_bins = 4
)
plot_bin_population(sce, best_bin(result), group_by_slot = "cell_type")
# Plot populations
sce <- assign_pseudotime_bins(
sce,
pseudotime_slot = "pseudotime", n_bins = 4
)
plot_bin_population(sce, best_bin(result), group_by_slot = "cell_type")
 # Getters
bulk_name(result)
#> [1] "B"
best_bin(result)
#> [1] 1
best_correlation(result)
#> [1] 1
top_2_distance(result)
#> [1] 0
confident_mapping(result)
#> [1] FALSE
mapping_history(result)
#> bin correlation lower_bound upper_bound
#> 1 1 1 1 1
#> 2 2 1 1 1
#> 3 3 1 1 1
#> 4 4 1 1 1
bootstrap_iterations(result)
#> [1] 200
# Getters
bulk_name(result)
#> [1] "B"
best_bin(result)
#> [1] 1
best_correlation(result)
#> [1] 1
top_2_distance(result)
#> [1] 0
confident_mapping(result)
#> [1] FALSE
mapping_history(result)
#> bin correlation lower_bound upper_bound
#> 1 1 1 1 1
#> 2 2 1 1 1
#> 3 3 1 1 1
#> 4 4 1 1 1
bootstrap_iterations(result)
#> [1] 200